Chemprop Application, Part I
Training the Chemprop Model
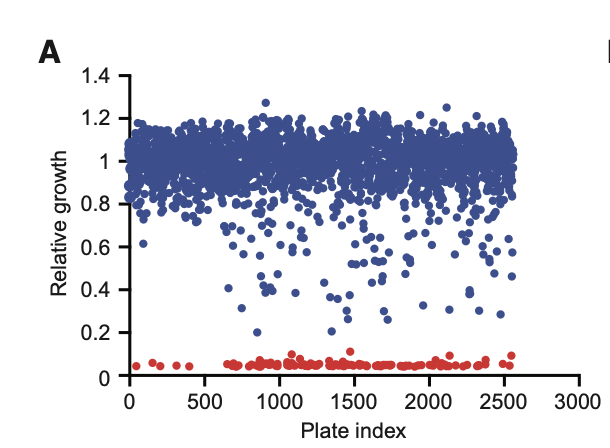
To begin, researchers trained Chemprop on a diverse dataset that included:
- 1,760 FDA-approved drugs
- 800 natural products from plants, animals, and microbes
- 2,560 total molecules tested for bacterial growth inhibition
Growth results were categorized by color:
Red = inhibitory activity, Blue = non-inhibitory.
After deduplication, 2,335 unique molecules remained.
Improving Model Performance
Due to the small size of the dataset, the researchers implemented several strategies to improve Chemprop’s performance. They engineered hundreds of molecular features to better represent chemical structures, applied hyperparameter optimization to fine-tune the model, and used ensembling to increase prediction stability. They also expanded the training set by incorporating data from external molecular databases. These steps helped make the model more accurate and effective when applied to new compounds.
External Validation on Drug Repurposing Hub
After training, the model was tested on the Drug Repurposing Hub, a publicly available library of bioactive compounds. This served as a validation step to evaluate generalization beyond the training data.

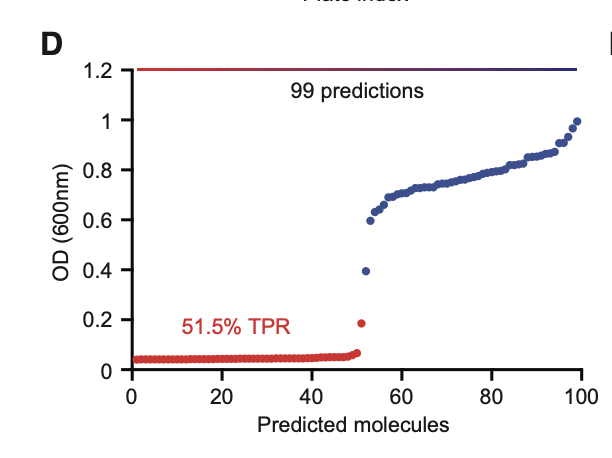
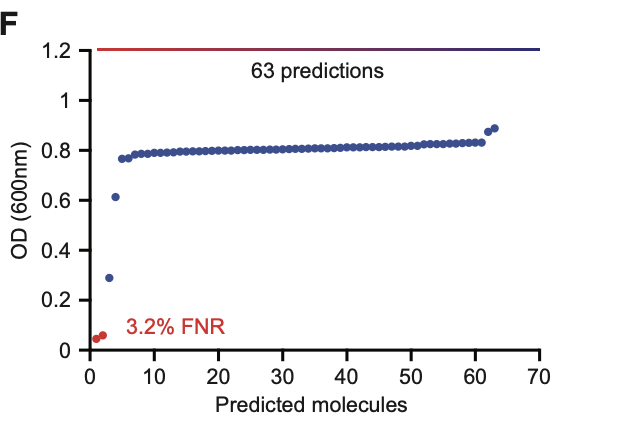
Chemprop’s predictions strongly aligned with observed biological activity.
There is a positive correlation between experimentally observed growth inhibition (red points) and the predicted rankings generated by the Chemprop model using D-MPNN.