Sequencing Technologies
There are many sequencing technologies in the field of DNA sequencing. Some of the most popular technologies currently include:
- Illumina - produces short read sequences (< 1kb), commonly used for whole genome sequencing, exomes, microRNA, and single-cell apps
- PacBio - produces long read sequences (~ 25 kb), used in the Revio sequencer at UVA
- Nanopore - produces “ultra-long” sequences (up to 1Mb)
- Hi-C - a crosslinking technique that captures interactions within genome
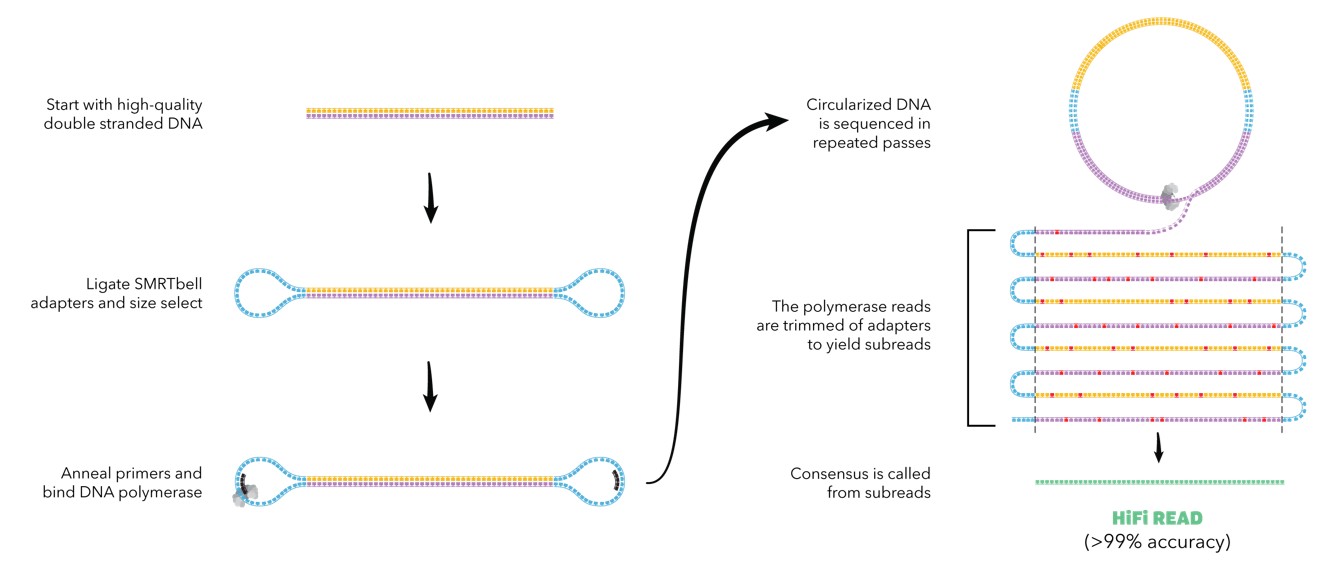
PacBio HiFi Reads
PacBio produces long reads typically around 25kb, with a 99.9% read quality (Q30), which is better than some short-read contigs. PacBio HiFi reads also distinguish repeats rather than spanning full repeats. However, their reads are limited in length and have lower throughput than Nanopore.
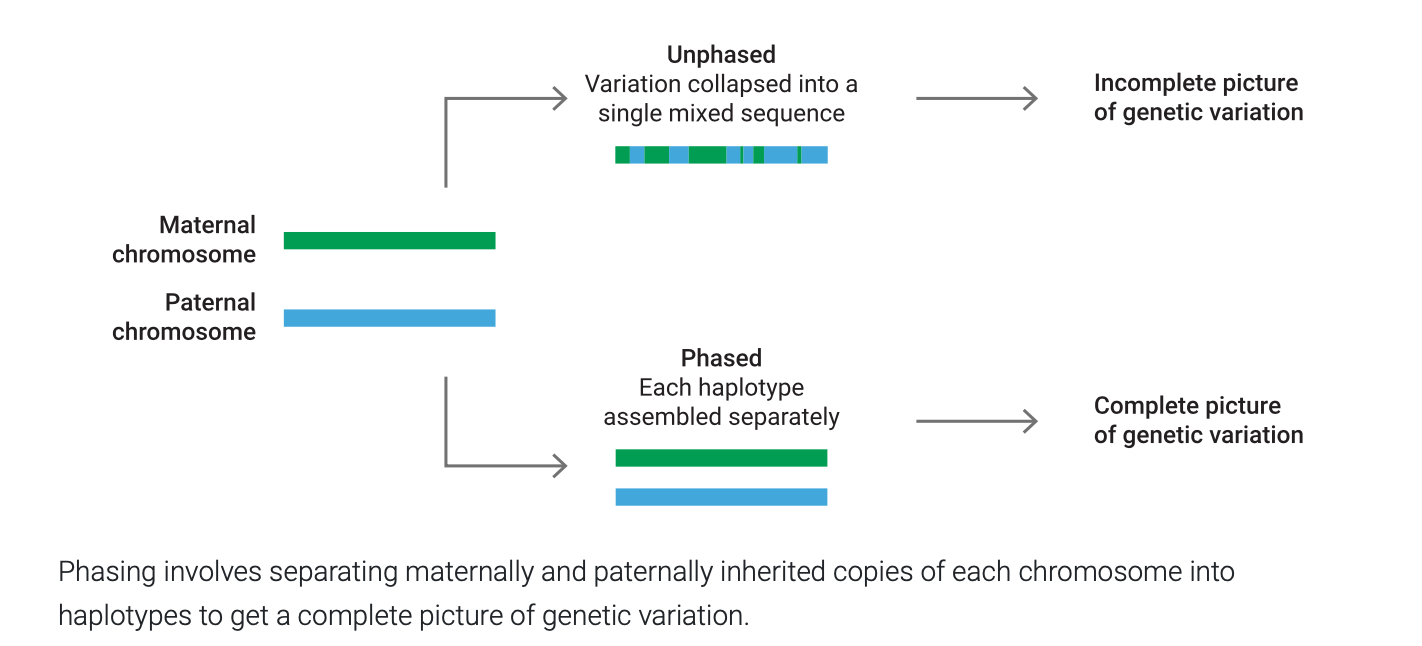
Phased Genome Assemblies
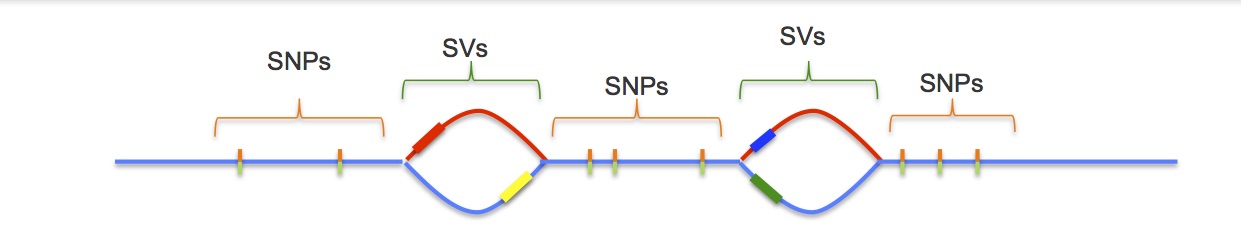
Standard assemblies produce unphased sequences, where the variation from two chromosomes is collapsed into a pseudo-haplotype (a single, mixed sequence). PacBio assemblies allow for the production of phased sequences, where the variation from two chromosomes is preserved in two separate sequences.
Nanopore Sequencing
Nanopore is able to produce “ultra-long” reads up to 4 Mb. Its accuracy is at about 95% read quality, and it is able to span repeat regions. The quality is limited compared to other technologies, and it has taken a while for Nanopore’s technology to reach where it is today.


Nanopore uses long reads and coverage to detect repeats.
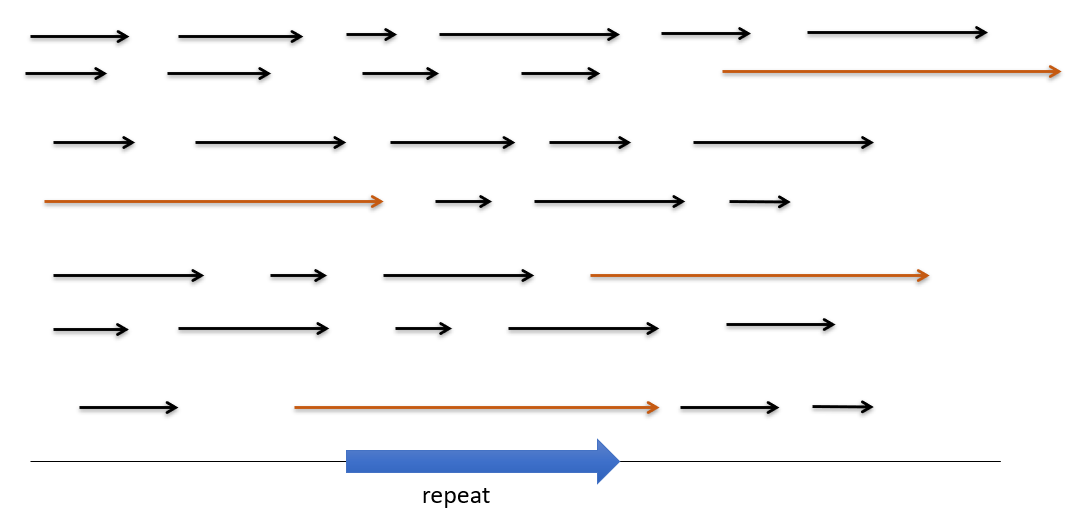
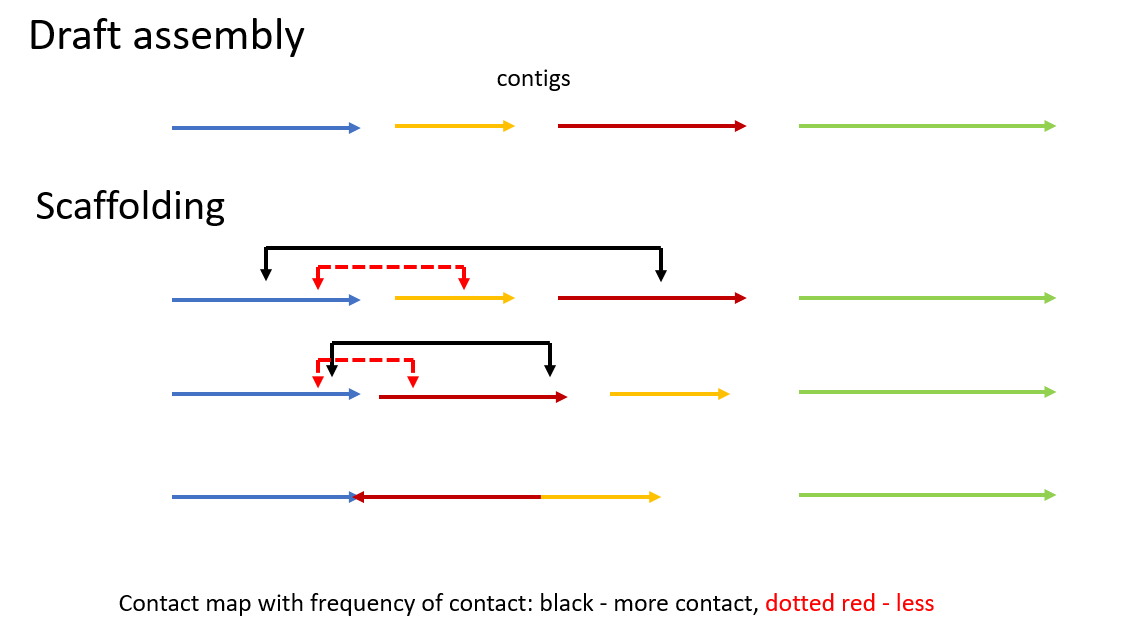
Hi-C
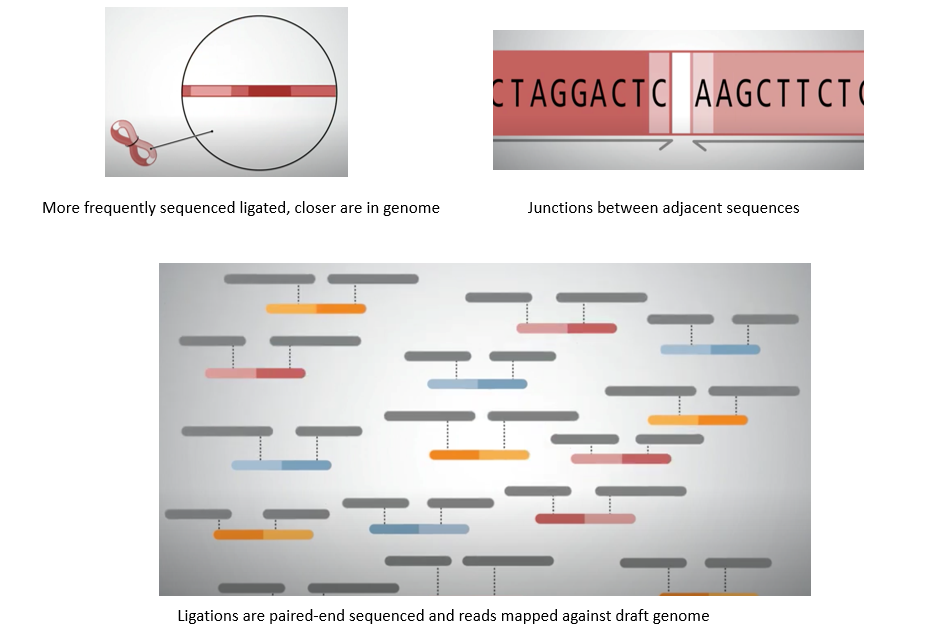
Hi-C is a genomic technique used to capture chromatin information. It involves detecting and analyzing the frequency of contacts between regions of DNA, in order to determine the correct order and orientation of the DNA segments. For example, segments with more contact are likely to be adjacent or closer to each other.
References
- Wenger et al. Nature Biotechnology 2019
- Chin et al. Nat Meth 2016
- PacBio: Sequencing 101 https://www.pacb.com/blog/ploidy-haplotypes-and-phasing/
- Shafin et al. Nature Biotechnology 2020
- How it Works: Proximo Hi-C Genome Scaffolding https://www.youtube.com/watch?v=-MxEw3IXUWU