RepeatMasker
Repetitive DNA sequences (biased nucleotide composition, tandem repeats, dispersed repeats, palindrome-hairpin structures, etc.) can prove to be issues when they are longer than the read length. RepeatMasker is a software tool helps tackle this problem by screening DNA sequences for interspersed repeats, and then masking/marking those repeats. The masking helps to prevent ambiguous alignments to regions of high similarity.
RepeatMasker uses two types of masking: soft and hard. Soft masking indicates masked regions by using lower-case letters. Hard masking (indicated with the -hardmask option) overwrites masked regions with a wildcard letter, using N for nucleotides or X for proteins.
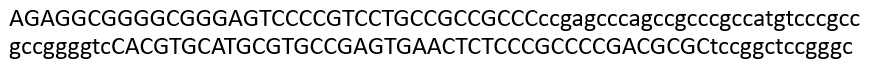
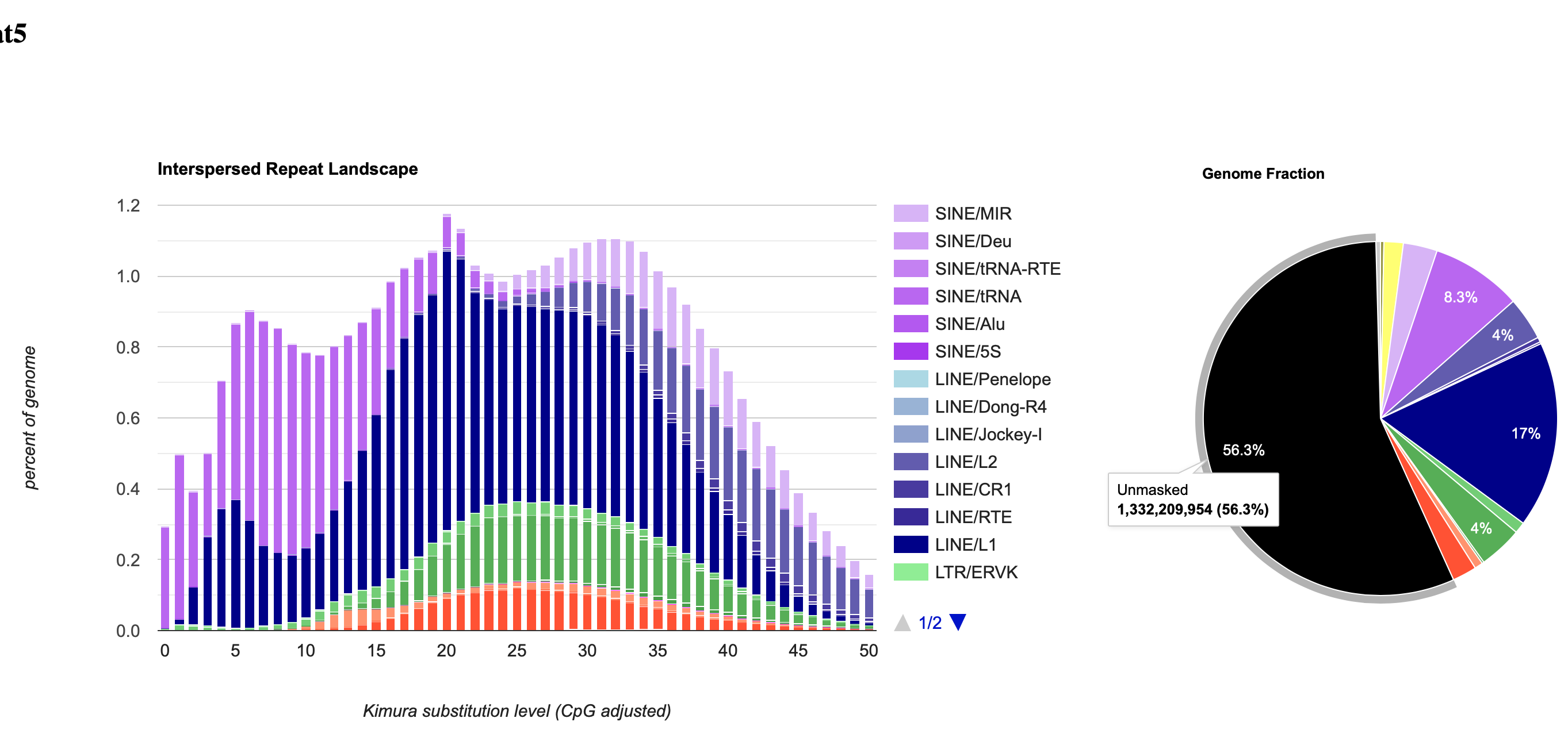
RepeatMasker is able to generate detailed annotations of the repeats in the DNA sequence, as well as a modified version of the DNA sequence in which all the annotated repeats have been masked (by default, replaced by Ns). Masking tools play a huge part in genomics research; for example, currently over 56% of the human genomic sequence is identified and masked by these programs.
Running RepeatMasker with a Slurm Script
Slurm is a resource manager that can be used to run your code for you. Below is a Slurm script called RepeatMasker_slurm_submit.sh.
#!/bin/bash
#SBATCH -A hpc_training # account name (--account)
#SBATCH -p standard # partition/queue (--partition)
#SBATCH --nodes=1 # number of nodes
#SBATCH --ntasks=1 # 1 task – how many copies of code to run
#SBATCH --cpus-per-task=4 # total cores per task – for multithreaded code
##SBATCH --mem=3200 # total memory (Mb) *Note ##comment
#SBATCH -t 01:00:00 # time limit: 1-hour
#SBATCH -J RepeatMasker-test # job name
#SBATCH -o RepeatMasker-test-%A.out # output file
#SBATCH -e RepeatMasker-test-%A.err # error file
#SBATCH --mail-user=dtriant@virginia.edu # where to send email alerts
#SBATCH --mail-type=ALL # receive email when starts/stops/fails
module purge # good practice to purge all modules
module load gcc/11.4.0
module load openmpi/4.1.4
module load repeatmasker/4.1.9
cd /project/rivanna-training/genomics-hpc/RepeatMasker
RepeatMasker genome_raw.fasta -lib Muco_library_EDTA.fasta -gff
After running the Slurm job, your output files will include masked sequence files, repeat statistics tables and .gff files.
For more info on RepeatMasker:
https://www.repeatmasker.org/
For info on interactive searching among commonly available genomes:
https://www.repeatmasker.org/cgi-bin/AnnotationRequest
For info on downloading raw annotation:
https://www.repeatmasker.org/genomicDatasets/RMGenomicDatasets.html