StringTie
StringTie aligns bulk RNA-Seq reads to a reference genome and estimates the abundance of a particular transcript or gene. It uses a metric called FPKM values, which stands for Fragments Per Kilobase of transcript per Million mapped reads. You can use the outputs from StringTie to estimate expression values.
Running StringTie with a Slurm Script
The stringtie_slurm_submit.sh script below is a Slurm script that will run StringTie. Slurm is a resource manager that can be used to run your code.
#!/bin/bash
#SBATCH -A hpc_training # account name (--account)
#SBATCH -p standard # partition/queue (--partition)
#SBATCH --nodes=1 # number of nodes
#SBATCH --ntasks=1 # 1 task – how many copies of code to run
#SBATCH --cpus-per-task=1 # total cores per task – for multithreaded code
##SBATCH --mem=3200 # total memory (Mb) *Note ##comment
#SBATCH -t 00:20:00 # time limit: 20 min
#SBATCH -J stringtie-test # job name
#SBATCH -o stringtie-test-%A.out # output file
#SBATCH -e stringtie-test-%A.err # error file
#SBATCH --mail-user=dtriant@virginia.edu # where to send email alerts
#SBATCH --mail-type=ALL # receive email when starts/stops/fails
module purge # good practice to purge all modules
module load stringtie/2.2.1
cd /project/rivanna-training/genomics-hpc/stringtie/tests_3 # working directory
stringtie --mix -G mix_guides.gff -o mix_reads_guided.out.gtf mix_short.bam mix_long.bam
# -- mix both short and long reads aligning
# -G reference annotation for guided alignment with short (1st) & long (2nd) read bam alignment files
# generates .gtf file with assembled transcripts and expression levels
Installing StringTie
If your desired module is not installed, you can submit a request via our ticket system (the wait time can vary depending on how popular the request is). You can also install and run StringTie locally.
git clone https://github.com/gpertea/stringtie
cd stringtie
make -j4 release
Look for a README file in the GitHub repository; the README often has download/installation instructions.
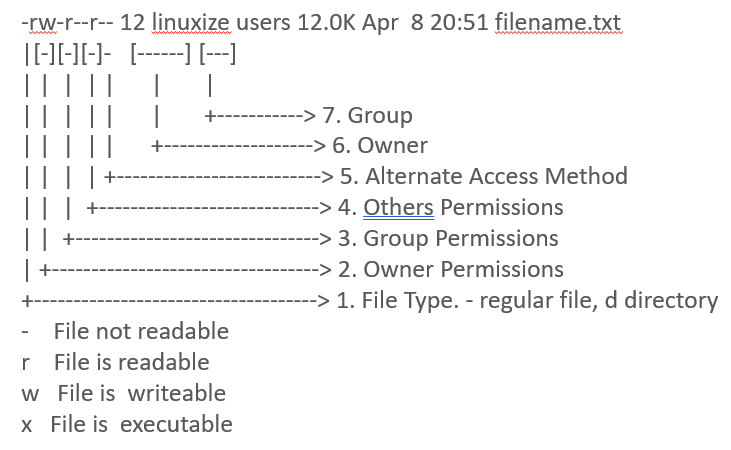
To check if your permissions are set to run StringTie, use the ls -lh command:
ls -lh stringtie
Running StringTie
Run ./run_test.sh to download and test sample data.
Test 1: Short reads
../stringtie -o short_reads.out.gtf short_reads.bam
Test 2: Short reads and super-reads
../stringtie -o short_reads_and_superreads.out.gtf short_reads_and_superreads.bam
Test 3: Short reads with annotation guides
../stringtie -G mix_guides.gff -o short_guided.out.gtf mix_short.bam
Test 4: Long reads
../stringtie -L -o long_reads.out.gtf long_reads.bam
Test 5: Long reads with annotation guides
../stringtie -L -G human-chr19_P.gff -o long_reads_guided.out.gtf long_reads.bam
Test 6: Mixed reads
../stringtie --mix -o mix_reads.out.gtf mix_short.bam mix_long.bam
Test 7: Mixed reads with annotation guides
../stringtie --mix -G mix_guides.gff -o mix_reads_guided.out.gtf mix_short.bam mix_long.bam
Test 8: Short reads with -N
../stringtie -N -G mix_guides.gff -o mix_short_N_guided.out.gtf mix_short.bam
Test 9: Short reads with --nasc
../stringtie --nasc -G mix_guides.gff -o mix_short_nasc_guided.out.gtf mix_short.bam
Running StringTie Example
Note your pathway when running StringTie: /project/rivanna-training/genomics-hpc/stringtie/stringtie.
We are going to run Test 7.
cd tests_3
stringtie --mix -G mix_guides.gff -o mix_reads_guided.out.gtf mix_short.bam mix_long.bam
In the command above, --mix sets the option to read short and long .bam files. The order of listed .bam files is important; the short .bam should be listed first, and the long .bam should be listed second. -G indicates that the annotation guide is a .gff file. The .gtf file is the output file.
For more information on StringTie: https://ccb.jhu.edu/software/stringtie/