CASP Competitions
CASP (Critical Assessment of Protein Structure Prediction) is a biennial competition for testing protein structure predictions started in 1994.
Participants are challenged to predict structures of proteins whose experimental structures have been recently solved but are not yet published or deposited in the PDB (Protein Data Bank).
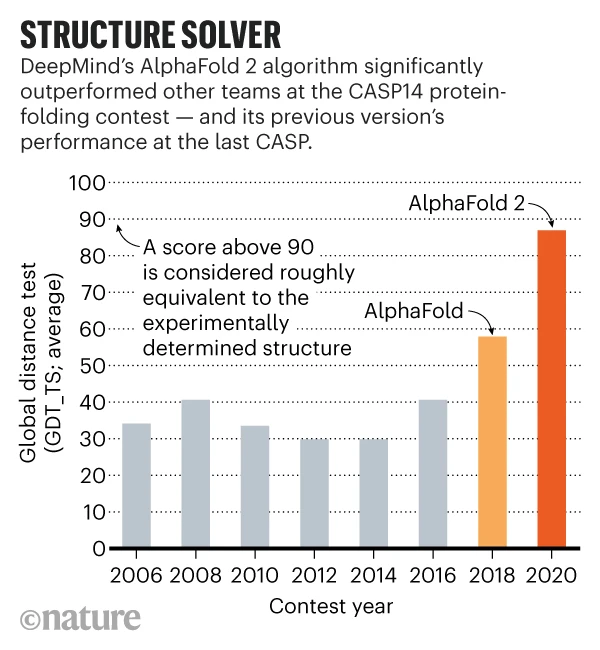
CASP uses the Global Distance Test - Total Score (GDT-TS) as a performance metric, which measures what percentage of α-carbons are within a threshold distance (in angstroms) of the experimental structure. The higher the GDT-TS score, the more accurate the model’s prediction is.
AlphaFold’s strong performance in CASP13 and CASP14 proved its ability to predict protein structures with near-experimental accuracy. This established AlphaFold as a game changer in the world of structural biology and computational science. Learn More