Alphafold Working Directory
AlphaFold is available as a module on Rivanna. Load it using:
module load apptainer/1.3.4 alphafold/2.3.0
View available AlphaFold versions:
module spider alphafold
More information on available versions
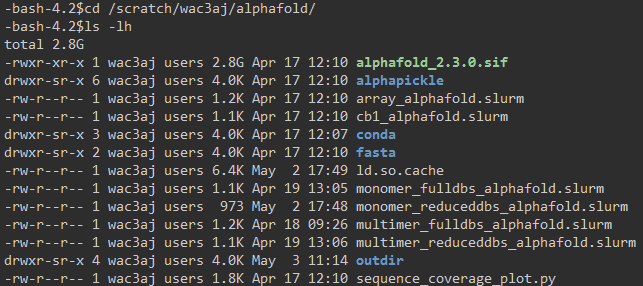
Directory Contents
The alphafold directory can contain the following:
-
SLURM job scripts (
*.slurm)
Several SLURM scripts are provided for submitting AlphaFold jobs for different use cases. -
fasta/
This subdirectory contains the FASTA files required for both monomer and multimer AlphaFold runs. -
conda/
This is a Conda environment that includes the necessary Python packages for running thealphapickleutility and thesequence_coverage_plot.pyscript. -
alphapickle/
This folder contains a GitHub package used to generate human-readable quality control plots and metric files (in.csvand.txtformat) as well as QC plots from the.pklresults generated by AlphaFold. -
sequence_coverage_plot.py
This is a custom script, modified from the VIBFold project, which generates sequence coverage plots using information from thefeatures.pklfile. -
outdir/
This directory is the destination for output files produced during AlphaFold runs.
View the files in your working directory:
cd alphafold
ls -lh